| Journal of Neurology Research, ISSN 1923-2845 print, 1923-2853 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Neurol Res and Elmer Press Inc |
| Journal website http://www.neurores.org |
Original Article
Volume 2, Number 5, October 2012, pages 204-210
Retinoic Acid Attenuates Cell Death and Reduces Tyrosine Hydroxylase Expression in Ethanol-Treated Human SH-SY5Y Neuroblastoma Cells
Khwanthana Grataitonga, Permphan Dharmasarojaa, b
aDepartment of Anatomy, Faculty of Science, Mahidol University, Rama VI Road, Ratchathewi, Bangkok 10400, Thailand
bCorresponding author: Permphan Dharmasaroja, Department of Anatomy, Faculty of Science, Mahidol University, Rama VI Road, Ratchathewi, Bangkok 10400, Thailand
Manuscript accepted for publication October 1, 2012
Short title: Retinoic Acid on Cell Death
doi: https://doi.org/10.4021/jnr147w
| Abstract | ▴Top |
Background: Retinoic acid (RA) plays an important role in embryonic development and central nervous system function, and has been shown to exert anti-apoptotic effects in various cells. In contrast, ethanol probably exerts its toxicity via pro-apoptotic effects. Here, we investigated the effects of all-trans RA on ethanol-induced cell death in human dopaminergic SH-SY5Y neuroblastoma cells.
Methods: Cell viability of SH-SY5Y cells exposed to 25, 50, 100, and 200 mM ethanol for 72 hours, with or without 10 µM RA, was measured using an MTT assay. Expression of p53, Bcl, and Bax mRNA levels in untreated SH-SY5Y cells and cells exposed to 200 mM ethanol, 10 µM RA, or both for 24 hours was analyzed using quantitative real-time RT-PCR. Tyrosine hydroxylase (TH) of SH-SY5Y cells exposed to 25, 50, 100, and 200 mM ethanol for 72 hours, with or without 10 µM RA, was measured using western blotting analysis.
Results: The effect of ethanol on cell viability was dose-dependent, and was accompanied by the significant reduction of p53 and Bax mRNA expression, including Bax/Bcl-2 ratio. Western blotting analysis showed that 72 hours of treatment with 200 mM ethanol significantly increased TH expression, but the expression was significantly decreased when cells were co-cultured with 10 µM RA and 200 mM ethanol.
Conclusions: All-trans RA could protect against apoptosis via a p53-dependent pathway and reduce the biochemical adaptation of ethanol-treated SH-SY5Y cells.
Keywords: Apoptosis; Ethanol; Retinoic acid; SH-SY5Y cells; Tyrosine hydroxylase
| Introduction | ▴Top |
In the presence of all-trans retinoic acid (RA), an active form of vitamin A, human SH-SY5Y neuroblastoma cells can differentiate into neuron-like cells, which exhibit long dendritic processes and express neuron-specific markers [1-3]. RA also plays an important role in embryonic development and central nervous system function. In contrast, ethanol inhibits neuronal differentiation and acts as a teratogen by causing brain abnormalities in fetal alcohol spectrum disorders [4]. The interaction between RA and ethanol has been widely investigated [5]. Previous studies have suggested potential mechanisms of toxicity by which ethanol might exert pro-apoptotic events in neuronal cells. Exposure to ethanol leads to a decrease in anti-apoptotic genes or proteins and an increase in pro-apoptotic genes or proteins in various neuronal cells, including Purkinje cells and neural crest cells [6-8]. RA has been shown to exert anti-apoptotic effects in various cells, including a human neuroblastoma cell line [9-11].
It is presumed that tyrosine hydroxylase (TH), the enzyme required for catecholamine synthesis, is constitutively expressed at a very low level in SH-SY5Y neuroblastoma cells [12]. SH-SY5Y cells differentiate into neuron-like cells in the presence of 10 µM all-trans RA [2]. TH, as an indicator of functional differentiation, is significantly increased in SH-SY5Y cells after three-day treatment with 10 µM all-trans RA. [12] Up-regulation of TH levels has also been reported as a hallmark of biochemical adaptations to ethanol exposure [13, 14].
The present paper aimed to investigate whether the anti-apoptotic potential of all-trans RA could prevent or attenuate the apoptotic effects of ethanol, thereby preventing the functional adaptation of ethanol-treated SH-SY5Y cells. Gaining a better understanding of how the ethanol effects could be prevented by RA might lead to developments of therapeutic or prophylactic treatment of ethanol-induced brain injuries.
| Materials and Methods | ▴Top |
Cell cultures
SH-SY5Y human neuroblastoma cells (passage 35) were maintained at 37 °C in a humidified atmosphere containing 5% CO2. Cells were grown in a 1:1 mixture of Eagle’s Minimum Essential Medium and Nutriebt Mixture Ham’s F12 medium, supplemented with 10% heat-activated fetal bovine serum (FBS). The medium contains 1 mM sodium pyruvate, 0.1 mM non-essential amino acid, 1.5 g/L sodium bicarbonate, 100 U/mL penicillin, and 100 µg/mL streptomycin. Cells were seeded at an initial density of 104 cells/cm2 in culture dishes. All-trans RA (Sigma-Aldrich Pte. Ltd., Singapore) dissolved in dimethylsulfoxide (DMSO; Fisher Scientific, Fair Lawn, NJ) was added a day after plating at a final concentration of 10 µM in MEM with 10% FBS.
Cell viability measurement by MTT assay
The 3-(4, 5-dimethylthaiazol-2-yl) 2, 5-diphenyl-tetrezolium bromide (MTT; Sigma-Aldrich Pte. Ltd., Singapore) was dissolved in Hank’s balanced salt solution (Sigma-Aldrich Pte. Ltd., Singapore), filtered, protected from light, and stored at 4 °C. SH-SY5Y cells were plated at density of 4 × 104 cells/well in 96-well tissue culture plates and allowed to grow for 24 hours.All-trans RA dissolved in DMSO was added a day after plating at a final concentration of 10 µM in MEM with 10% FBS. In a time-dependent experiment, SH-SY5Y cells were additionally exposed to 200 mM ethanol for 24, 48, and 72 hours. In a dose-dependent experiment and co-treatment of ethanol and RA experiment, SH-SY5Y cells were additionally exposed to 25, 50, 100, or 200 mM ethanol for 72 hours and co-treated with 10 µM RA. Each treatment was performed in triplicate, and the assays were repeated in three independent experiments. SH-SY5Y cells were added with 5 mg/mL of MTT solution in culture medium, followed by incubation for 4 hours at 37 °C in a humidified atmosphere containing 5% CO2. Then the media were removed out and replaced with MTT solubilization solution to allow dissolution of formazan crystals. The absorbance of the samples was measured at 570 - 690 nm using a Biotek Synergy HT microplate reader with KC4 software.
Quantitative real-time reverse transcription polymerase chain reaction
Total mRNAs were extracted from untreated SH-SY5Y cells or cells exposed to 200 mM ethanol, 10 µM RA, or both for 24 hours using PARIS™ kit (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol. The purity of the RNAs samples was determined by the OD A260/A280 ratio with 1.8 or above using a spectrophotometer, 2 µg of total RNA were used for the reverse transcription using Masterscript RT-PCR System (5 PRIME, Gaithersburg, MD), according to the manufacturer’s instruction. cDNAs in each sample were amplified using the KAPA SYBR® FAST qPCR kit (Kapa Biosystems, Woburn, MA). Real-time PCR mixture was prepared in a volume of 20 µL, containing 10 µL of 1x KAPA SYBR® FAST qPCR master mix, 200 nM of forward and reverse primers, and nuclease-free water. 20 ng cDNA was used as PCR template. β-actin was used as a reference gene. The sequences of the primers for the qRT-PCR are as follows: p53: sense, 5′-GGAGGTTGTGAGGCGCTGG-3′; antisense, 5′-CACGCACCTCAAAGCTGTTC-3′; Bax: sense, 5′-TGCAGAGGATGATTGCTGAC-3′; antisense, 5′-GAGGACTCCAGCCACAAAGA-3′; Bcl-2: sense, 5′-CAGCTGCACCTGACG-3′; antisense, 5′-ATGCACCTACCCAGC-3′; TH: sense, 5′-GCCGTGCTAAACCTGCTCTT-3′; antisense, 5′-GTCTCAAACACCTTCACAGCTC-3′; β-actin: sense, 5′-CATGTACGTTGCTATCCAGGC-3′; antisense, 5′-CTCCTTAATGTCACGCACGAT-3′. The reaction was performed in the Applied Biosystems 7500 Real-time PCR system, with the PCR cycling conditions as follows: step 1 enzyme activation, 95°C for 20 sec-3 min; step 2 amplification, 40 cycles at 95 °C for 1 - 3 sec; step 3 annealing step, 60 °C for 1 min. Melting curve analysis was performed from 65 to 94 °C at the rate of 0.1°C/seconds after PCR to ensure amplification specificity. The transcript levels of the target genes were determined by the comparative threshold cycle (Ct) method, and Ct number for each PCR was defined using the second derivative maximum algorithm that identifies the first turning point of the fluorescence curve, as determined by the ABI 7500 software, version 2.0.
Western blot analysis
SH-SY5Y cells treated as indicated in the result section were placed on ice, and washed three times with PBS. Cells were lysed with RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% triton X-100, 0.1% SDS, 1% sodium deoxycholate, 5 mM EDTA, 30 mM Na2HPO4, 50 mM NaF). Cellular debris was precipitated by centrifugation at 12,000 rpm and 4 °C for 15 min. The supernatants were collected and protein concentration was determined using the Bradford reagent (Bio-Rad Laboratories, Hercules, CA), 20 µg for each sample were separated by 12.5% SDS-polyacrylamide gels. After electrophoresis, proteins were blotted onto nitrocellulose membranes. To determine equal loading and blotting, gels were stained with Coomassie Brilliant Blue and membranes were stained with Ponceau S. After staining, membranes were blocked with 5% skim milk in TBS-T at 4 °C overnight. Then, membranes were incubated with 1:1000 primary rabbit polyclonal antibody to TH (Cell Signal Technology, Danvers, MA) in TBS-T at 4 °C overnight. After incubation with the primary antibody, membranes were rinsed three times with TBS-T and incubated at room temperature for 1 hour with 1:5,000 horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (Chemicon International Inc., Billerica, MA). The immunoreactive proteins were detected by enhanced chemiluminescent (ECL) reagents (Thermo Fisher Scientific, Rockford, IL). Quantification of western blots was performed by densitometric analysis using Scion Image software.
Statistical analyses
Statistical analyses were performed using GraphPad Prism software for Windows (GraphPad Software, San diego, CA). All results were shown as mean ± standard error of mean (SEM). Differences in the time course profiles of TH mRNA expression between various treatment groups were calculated using one-way analysis of variance (ANOVA) with Tukey multiple comparison tests for overall statistical significance, while differences at a single time point were calculated by two-tailed, unpaired t-test. Results were considered significant for P < 0.05.
| Results | ▴Top |
RA reduced ethanol-induced cell death
Using an MTT assay, SH-SY5Y cells exposed to ethanol at 100 and 200 mM for 72 hours had their respective cell viability of 84 ± 4 (P < 0.05) and 68 ± 1% (P < 0.001), which were significantly lower than unexposed control (Fig. 1A). The effect of ethanol on cell viability was dose-dependent. Cells treated with 200 mM ethanol for 72 hours had significantly lower viability than those treated with 100 mM ethanol (P < 0.05). In the presence of 10 µM RA, which was added to the culture media at day 0, there were no significant changes in cell viability of SH-SH5Y cells exposed to 25, 50, 100, and 200 mM ethanol for 72 hours when compared with unexposed cells with or without RA (Fig. 1B).
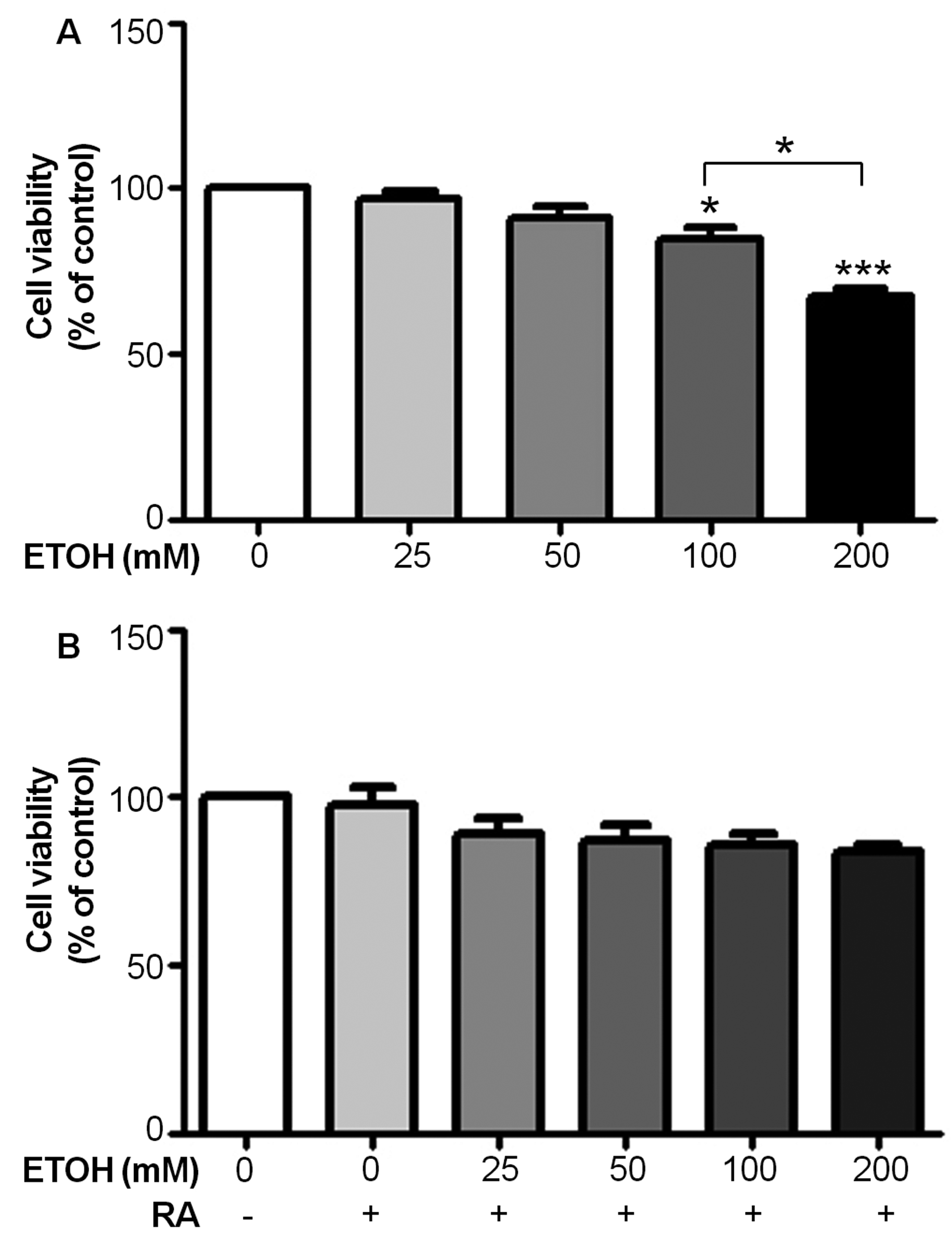 Click for large image | Figure 1. Retinoic acid (RA) reduces ethanol-induced cell death as assessed by an MTT assay. (A) Cell viability of SH-SY5Y cells after exposure to various concentrations of ethanol for 72 hours. Data are expressed as the mean of three replicate ± SEM from three independent experiments (*P < 0.05 and***P < 0.001). (B) Cell viability of SH-SY5Y cells after co-treatment with 10 µM RA and various concentrations of ethanol for 72 hours. Data are expressed as the mean of three replicate ± SEM from three independent experiments. |
RA reduced p53 and Bax/Bcl-2 mRNA expression in ethanol-treated SH-SY5Y cells
Using real-time quantitative RT-PCR analysis, the expression of p53 mRNA was significantly increased in 200 mM ethanol-treated SH-SY5Y cells after 24 hours exposure when compared to unexposed cells (P < 0.01; Fig. 2A). Co-treatment of 10 µM RA and 200 mM ethanol significantly reduced p53 expression when compared with cells cultured in ethanol alone (P < 0.05). Treatment with 200 mM ethanol also significantly increased the expression of pro-apoptotic Bax (P < 0.05; Fig. 2B) and significantly decreased the expression of anti-apoptotic Bcl-2 mRNA (P <0.05; Fig. 2C) when compared to untreated cells. As a result, Bax/Bcl-2 expression was significantly increased in SH-SY5Y cells treated with 200 mM ethanol for 24 hours (P < 0.001; Fig. 2D), whereas Bax/Bcl-2 expression was significantly lower in cells co-cultured in 10 µM RA and 200 mM ethanol (P < 0.01). These findings suggest that the pro-survival effect of RA in ethanol-induced apoptotic SH-SY5Y cells is mediated by a change in p53-dependent signaling.
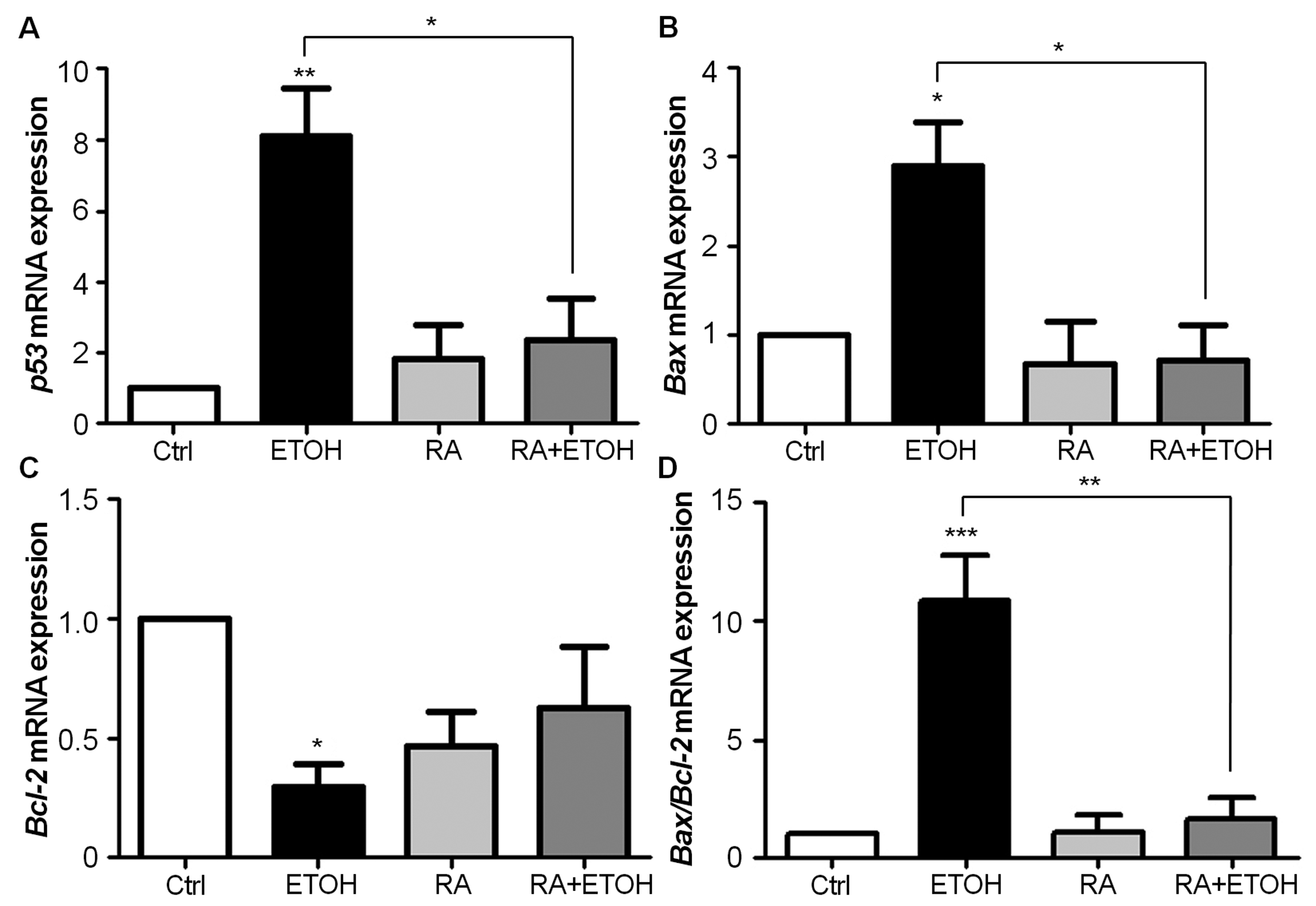 Click for large image | Figure 2. (A-D) RA reduces p53 and Bax/Bcl-2 mRNA expression in ethanol-treated SH-SY5Y cells. Cells were incubated with 200 mM ethanol, 10 µM RA, and 200 mM ethanol plus 10 µM RA for 24 hours in each experiment. The expression of p53 (A), Bax (B), Bcl-2 (C), and Bax/Bcl-2 ratio (D) was analyzed by quantitative real-time RT-PCR, and relatively compared to their respective untreated control. The expression levels, standardized by the level of β-actin, were shown as folds of expression. Each bar represents the mean of three replicate ± SEM from three independent experiments (*P < 0.05, ** P < 0.01, and *** P < 0.001). |
RA attenuated tyrosine hydroxylase expression in ethanol-treated SH-SY5Y cells
The incubation of SH-SY5Y cells in ethanol for 72 hours increased tyrosine hydroxylase (TH) protein expression in a dose-dependent manner (Fig. 3A). Significant increases in TH expression were observed when cells were exposed to 200 mM ethanol when compared to unexposed cells and cells exposed to lesser concentrations of ethanol (P < 0.001). As shown inFigure 3B, the incubation of SH-SY5Y cells in 200 mM ethanol for 72 hours significantly increased TH protein levels (P < 0.01) compared to unexposed cells; and cells cultured in 10 µM RA and 200 mM ethanol showed significantly decreased TH protein levels (P < 0.01) when compared to ethanol-treated cells, suggesting that RA reduced the ethanol-induced functional changes in SH-SY5Y cells.
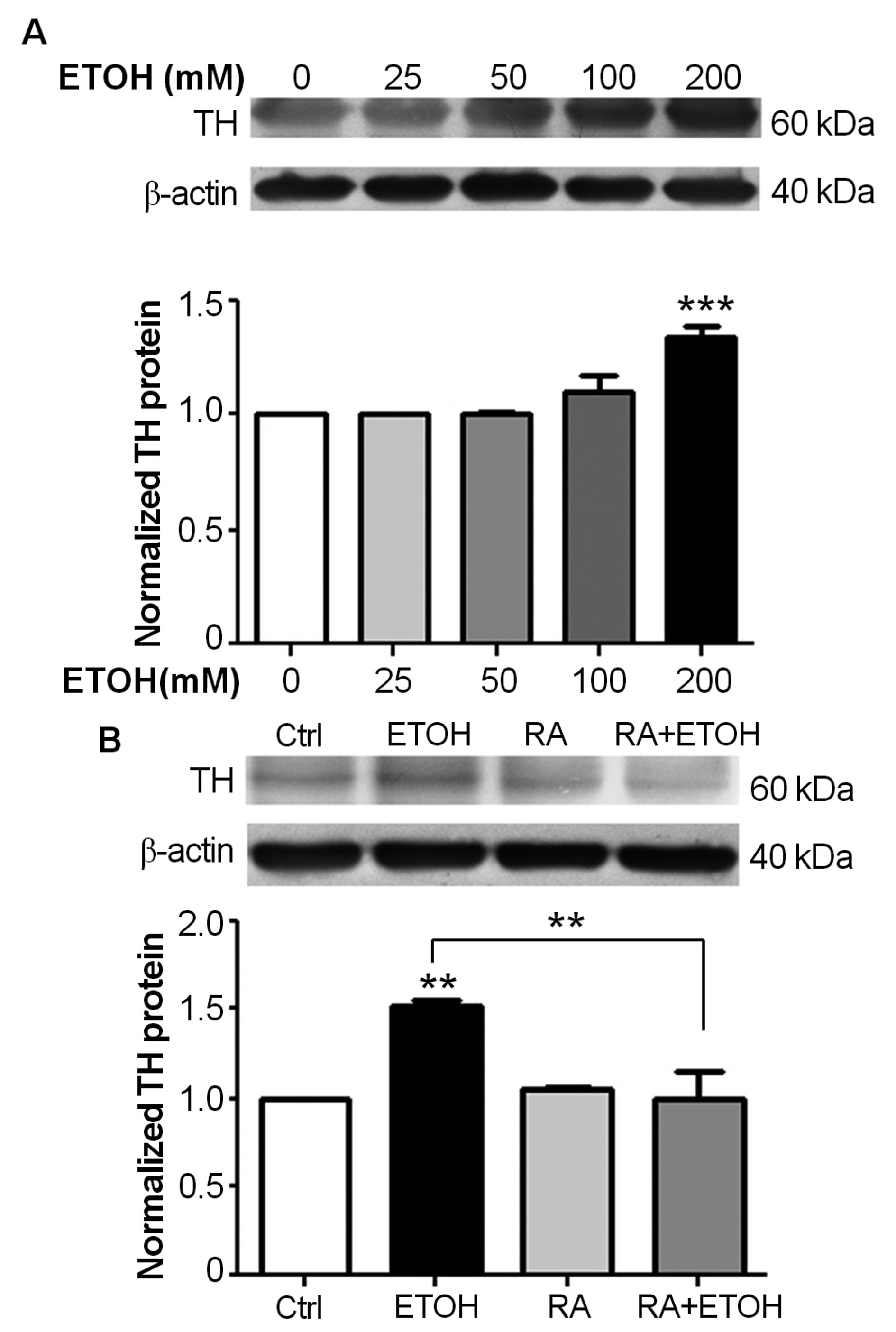 Click for large image | Figure 3. Retinoic acid (RA) reduces tyrosine hydroxylase (TH) protein expression. Total proteins were separated by 12.5% SDS-PAGE and were analyzed by immunoblotting with anti-TH antibody and anti-actin antibody. The expression levels of TH were relatively compared to untreated control, and were shown as folds of expression. Each bar represents the mean of three replicate ± SEM from three independent experiments. (A) Ethanol induces TH protein expression in a dose-dependent manner. Proteins were isolated from SH-SY5Y cells after exposure to 25, 50, 100, and 200 mM ethanol for 72 hours (***P < 0.001). (B) RA reduces TH protein expression in ethanol-treated SH-SY5Y cells. Total proteins were isolated from SH-SY5Y cells after 72 hours exposure to 200 mM ethanol, 10 µM RA, and 200 mM ethanol plus 10 µM RA (**P < 0.01). |
| Discussion | ▴Top |
Experiments using primary neural cultures and neuronal cell lines have shown that ethanol induces cell death via apoptosis [6, 7]. The modulatory role of RA in apoptosis is controversial. In the present study, treatment of 10 µM all-trans RA reduced ethanol-induced cell death, and reversed mRNA expression levels of p53, Bcl, and BAX in human SH-SY5Y neuroblastoma cells. We also showed that the deleterious effects of ethanol on TH protein and mRNA expression were significantly attenuated by RA.
Previous studies have shown that the mechanism of ethanol-induced apoptosis is mediated via a p53-dependent pathway [15, 16]. The connection between p53 and Bcl-2 family members has been established [17]. Pro-apoptotic Bax, directly induced by p53, can overcome the anti-apoptotic effect of Bcl-2, whereas p53 can directly inhibit Bcl-2. In the present study, treatment with 200 mM ethanol significantly increased the expression of p53. As a result, increased p53 induced Bax expression and inhibited Bcl-2 expression, leading to the apoptosis of SH-SY5Y cells. These ethanol effects were significantly reduced when cells were cultured in the presence of 10 µM all-trans RA. The effects of RA on the expression of Bax and Bcl-2 have been demonstrated in previously stress- or apoptosis-induced cells [8, 9, 18, 19]. Our study showed that the exposure of SH-SY5Y cells to 10 µM RA alone for 24 hours did not cause a significant change in the expression of p53, Bax, and Bcl-2 mRNA when compared to untreated controls. However, when cells were cultured in the presence of both 10 µM RA and 200 mM ethanol, a significant shift in the expression of p53, Bax, and Bax/Bcl-2 ratio were demonstrated. These results suggest that in ethanol-induced apoptotic SH-SY5Y cells, the pro-survival effect of RA might be mediated by a change in p53-dependent signaling.
The present study showed that ethanol increased TH protein expression in a dose- and time-dependent manner. A significant change was observed at 200 mM ethanol and following 72 hours of exposure. A previous study suggested that ethanol-induced expression of TH in SH-SY5Y cells is probably mediated by the formation of the TH-HSP90 (heat shock protein 90) complex via cyclic AMP-dependent protein kinase A (PKA) [14]. TH is involved in the biosynthesis of catecholaminergic neurotransmitters, including dopamine. Dopaminergic neurons can adaptively increase TH protein levels in response to ethanol exposure and this mechanism may be related to drug addiction [13]. Treatment of SH-SY5Y cells with 10 µM RA alone for 72 hours increases TH immunoreactivity, as demonstrated in our study and elsewhere [12]. These results indicate that both ethanol and RA cause functional changes in SH-SY5Y cells, which constitutively express TH at a very low level, into dopaminergic neuron-like cells.
The present study yielded the novel result that, in the presence of 10 µM RA, the expressions of TH protein and mRNA were significantly reduced in 200 mM ethanol-treated SH-SY5Y cells, suggesting that RA prevented the ethanol-induced functional changes in SH-SY5Y cells. A previous study showed that glial cell line-derived neurotrophic factor (GDNF) reverses biochemical adaptation of SH-SY5Y cells to ethanol exposure by inhibiting the interaction between TH and HSP90, leading to the down-regulation of TH protein stability [14]. Evidence in developing rat sympathetic neurons showed that RA acts through an α-type of nuclear retinoic acid receptor to induce GDNF responsiveness [20]. Thus, a mechanism by which RA reduces TH expression in ethanol-treated cells might be RA-mediated GDNF responsiveness, resulting in an impairment of TH-HSP90 association. Further investigation is required to address this hypothesis.
The reversal of ethanol effects by all-trans RA has been reported. In quail embryo cultures, the addition of RA reverses ethanol-induced cardiovascular abnormalities [21]. The adverse effects of ethanol on neurite extension in human LA-N-5 neuroblastoma cells is also prevented by RA, likely by increasing growth-associated protein 43 (GAP43/B50) and decreasing N-myc and c-myc protein levels [22]. Activation of myc increases p53 levels in neuroblastoma cells and leads to apoptosis [23]. Thus, a possible mechanism of RA in the reversal of ethanol effects might be mediated via decreasing myc levels, leading to reduced p53 activation and the prevention of apoptosis. p53 inactivation in response to GDNF has been reported, and may also contribute to the anti-apoptotic role of RA [24].
In conclusion, we demonstrated that all-trans RA could protect against ethanol-induced cell death and reduce the functional adaptation of ethanol-induced TH expression in SH-SY5Y cells. The effect of RA is likely mediated via a p53-dependent pathway. The results might shed light in clinical applications in fetal alcohol spectrum disorders and ethanol-related brain injuries.
Financial Disclosures
This study was supported in part by a grant from Department of Anatomy, Faculty of Science, Mahidol University, to Permphan Dharmasaroja.
Source of Grant Support
This study was supported in part by Department of Anatomy, Faculty of Science, Mahidol University. There was no conflict of interest.
| References | ▴Top |
- Pahlman S, Ruusala AI, Abrahamsson L, Mattsson ME, Esscher T. Retinoic acid-induced differentiation of cultured human neuroblastoma cells: a comparison with phorbolester-induced differentiation. Cell Differ. 1984;14(2):135-144.
doi pubmed - Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Cena V, Gallego C, et al. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem. 2000;75(3):991-1003.
doi pubmed - Miloso M, Villa D, Crimi M, Galbiati S, Donzelli E, Nicolini G, Tredici G. Retinoic acid-induced neuritogenesis of human neuroblastoma SH-SY5Y cells is ERK independent and PKC dependent. J Neurosci Res. 2004;75(2):241-252.
doi pubmed - Duester G. A hypothetical mechanism for fetal alcohol syndrome involving ethanol inhibition of retinoic acid synthesis at the alcohol dehydrogenase step. Alcohol Clin Exp Res. 1991;15(3):568-572.
doi pubmed - Wolf G. Tissue-specific increases in endogenous all-trans retinoic acid: possible contributing factor in ethanol toxicity. Nutr Rev. 2010;68(11):689-692.
doi pubmed - Heaton MB, Madorsky I, Paiva M, Mayer J. Influence of ethanol on neonatal cerebellum of BDNF gene-deleted animals: analyses of effects on Purkinje cells, apoptosis-related proteins, and endogenous antioxidants. J Neurobiol. 2002;51(2):160-176.
doi pubmed - Siler-Marsiglio KI, Paiva M, Madorsky I, Pan Q, Shaw G, Heaton MB. Functional mechanisms of apoptosis-related proteins in neonatal rat cerebellum are differentially influenced by ethanol at postnatal days 4 and 7. J Neurosci Res. 2005;81(5):632-643.
doi pubmed - Wentzel P, Eriksson UJ. Altered gene expression in neural crest cells exposed to ethanol in vitro. Brain Res. 2009;1305 Suppl:S50-60.
pubmed - Lombet A, Zujovic V, Kandouz M, Billardon C, Carvajal-Gonzalez S, Gompel A, Rostene W. Resistance to induced apoptosis in the human neuroblastoma cell line SK-N-SH in relation to neuronal differentiation. Role of Bcl-2 protein family. Eur J Biochem. 2001;268(5):1352-1362.
doi pubmed - Kholodenko R, Kholodenko I, Sorokin V, Tolmazova A, Sazonova O, Buzdin A. Anti-apoptotic effect of retinoic acid on retinal progenitor cells mediated by a protein kinase A-dependent mechanism. Cell Res. 2007;17(2):151-162.
doi pubmed - Sakamoto K, Hiraiwa M, Saito M, Nakahara T, Sato Y, Nagao T, Ishii K. Protective effect of all-trans retinoic acid on NMDA-induced neuronal cell death in rat retina. Eur J Pharmacol. 2010;635(1-3):56-61.
pubmed - Kume T, Kawato Y, Osakada F, Izumi Y, Katsuki H, Nakagawa T, Kaneko S, et al. Dibutyryl cyclic AMP induces differentiation of human neuroblastoma SH-SY5Y cells into a noradrenergic phenotype. Neurosci Lett. 2008;443(3):199-203.
doi pubmed - Ortiz J, Fitzgerald LW, Charlton M, Lane S, Trevisan L, Guitart X, Shoemaker W, et al. Biochemical actions of chronic ethanol exposure in the mesolimbic dopamine system. Synapse. 1995;21(4):289-298.
doi pubmed - He DY, Ron D. Glial cell line-derived neurotrophic factor reverses ethanol-mediated increases in tyrosine hydroxylase immunoreactivity via altering the activity of heat shock protein 90. J Biol Chem. 2008;283(19):12811-12818.
doi pubmed - Ghosh AP, Walls KC, Klocke BJ, Toms R, Strasser A, Roth KA. The proapoptotic BH3-only, Bcl-2 family member, Puma is critical for acute ethanol-induced neuronal apoptosis. J Neuropathol Exp Neurol. 2009;68(7):747-756.
doi pubmed - Meng XF, Zou XJ, Peng B, Shi J, Guan XM, Zhang C. Inhibition of ethanol-induced toxicity by tanshinone IIA in PC12 cells. Acta Pharmacol Sin. 2006;27(6):659-664.
doi pubmed - Hemann MT, Lowe SW. The p53-Bcl-2 connection. Cell Death Differ. 2006;13(8):1256-1259.
doi pubmed - Choudhary R, Baker KM, Pan J. All-trans retinoic acid prevents angiotensin II- and mechanical stretch-induced reactive oxygen species generation and cardiomyocyte apoptosis. J Cell Physiol. 2008;215(1):172-181.
doi pubmed - Beveridge NJ, Tooney PA, Carroll AP, Tran N, Cairns MJ. Down-regulation of miR-17 family expression in response to retinoic acid induced neuronal differentiation. Cell Signal. 2009;21(12):1837-1845.
doi pubmed - Thang SH, Kobayashi M, Matsuoka I. Regulation of glial cell line-derived neurotrophic factor responsiveness in developing rat sympathetic neurons by retinoic acid and bone morphogenetic protein-2. J Neurosci. 2000;20(8):2917-2925.
pubmed - Twal WO, Zile MH. Retinoic acid reverses ethanol-induced cardiovascular abnormalities in quail embryos. Alcohol Clin Exp Res. 1997;21(6):1137-1143.
doi pubmed - Saunders DE, Hannigan JH, Zajac CS, Wappler NL. Reversal of alcohol's effects on neurite extension and on neuronal GAP43/B50, N-myc, and c-myc protein levels by retinoic acid. Brain Res Dev Brain Res. 1995;86(1-2):16-23.
pubmed - Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A, Shohet JM. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci U S A. 2005;102(3):731-736.
doi pubmed - Saifudeen Z, Dipp S, Stefkova J, Yao X, Lookabaugh S, El-Dahr SS. p53 regulates metanephric development. J Am Soc Nephrol. 2009;20(11):2328-2337.
doi pubmed
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Neurology Research is published by Elmer Press Inc.
