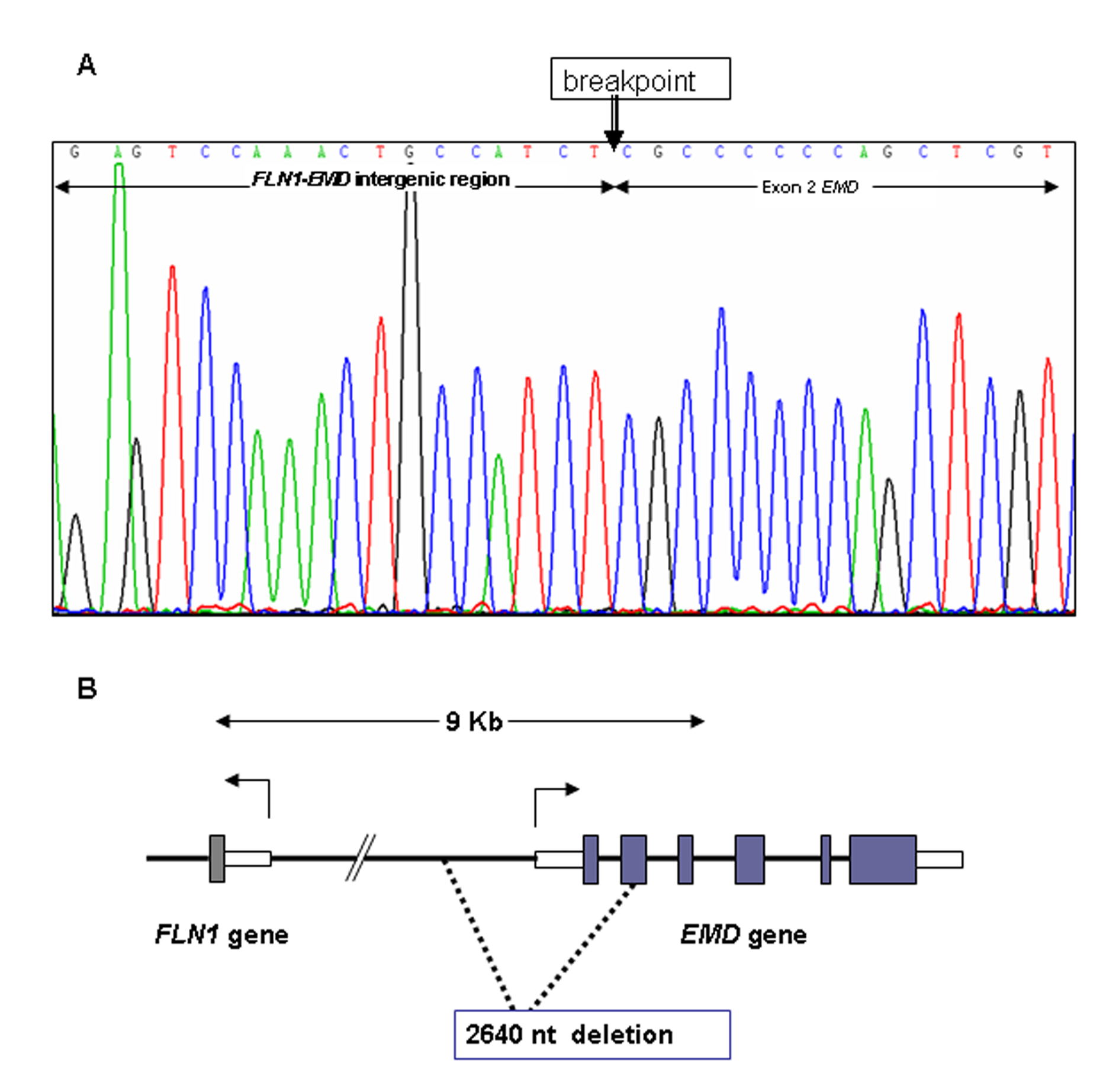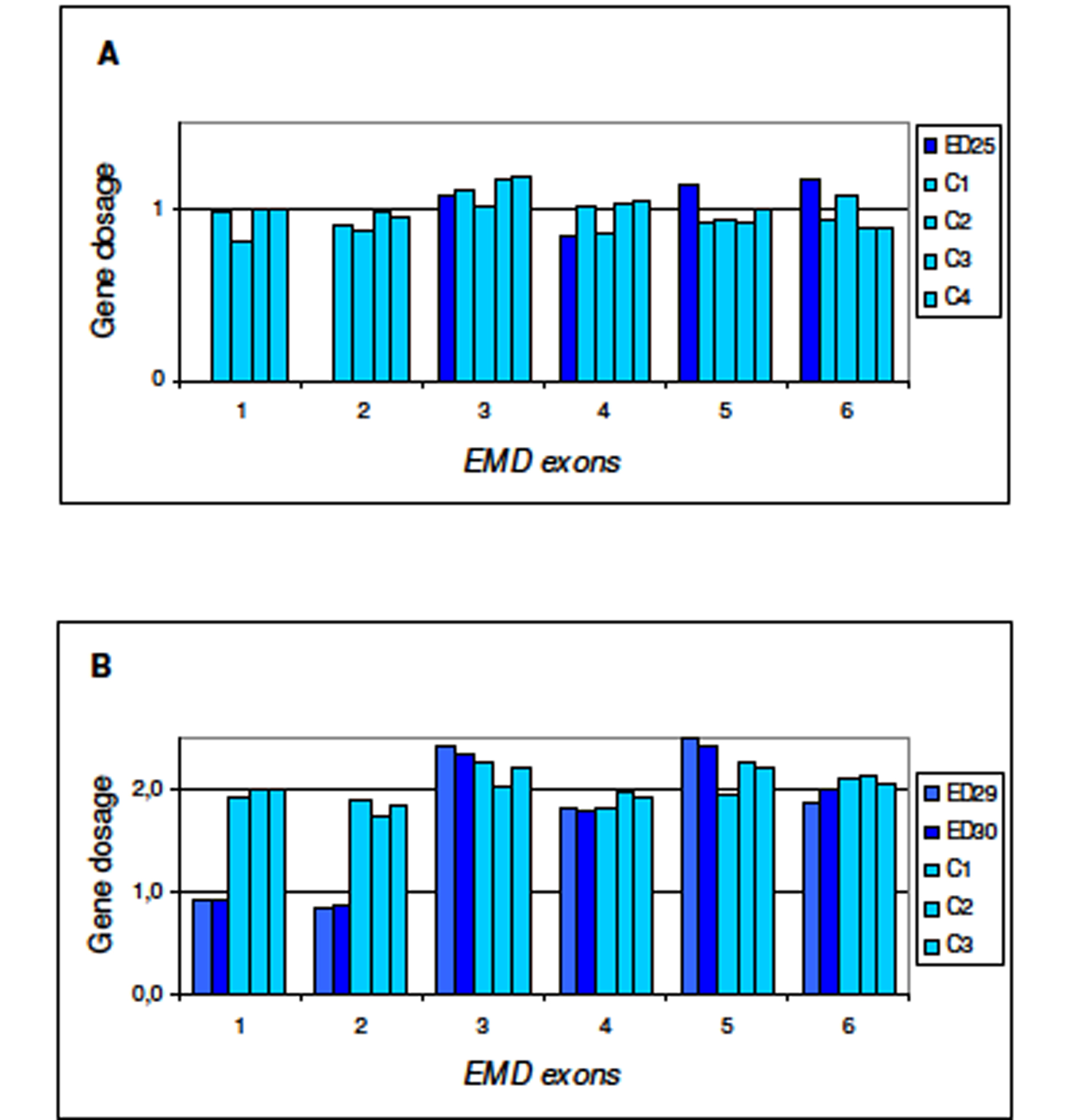
Figure 1. Multiplex Ligation-dependent Probe Amplification (MLPA) for dosage of EMD gene. Procedure for test performance was the one described by Shouten et al. [http://www.neurores.org/index.php/neurores/article/view/107/125 9]. For data analysis a global normalisation was achieved by dividing the peak area for each amplification product by the average peak area of all peaks in that sample. Normalised peak areas were compared to the results obtained for several control samples. In the final step, the results obtained were visualised using Excel graphics. In abscissa exons 1 to 6 of EMD gene and in ordinate gene dosage as deduced from peak areas. A) ED25 is the propositus; C1, C2, C3, and C4 are male controls. B) ED29 and ED30 are the mother and sister of the propositus, respectively; C1, C2, C3 are female controls.